
Saint-Antoine Hospital - Kourilsky Building – 5th floor
184, rue du Faubourg Saint-Antoine - 75012 Paris - France
1.Our Interests
Our team is interested in the pathophysiology of growth disorders involving the Insulin-like growth factors (IGFs) system. The IGF system is highly involved in cell metabolism, proliferation, differentiation and survival, particularly during development. IGF2 plays a major role in the control of fetal growth and has the particularity of being encoded by an imprinted gene. Parental genomic imprinting is an epigenetic process that results in the mono-allelic expression of certain genes according to their parental origin. This expression is under the control of Imprinting Control Region, ICR, which are differentially methylated regions between the two alleles.
The team is working on two rare, clinically and molecularly mirrored diseases secondary to the disruption of parental imprinting in the 11p15 region and affecting fetal growth: Silver-Russell syndrome (SRS, growth retardation) and Beckwith-Wiedmann syndrome (BWS, overgrowth with an increased risk of embryonic tumour development). We also reported the first French cohort of patients with Temple syndrome (TS14), another imprinting-related disease responsible for growth retardation and early metabolic and endocrine disorders, secondary to molecular abnormalities of the 14q32 region. These syndromes are secondary to genetic or epigenetic abnormalities (in particular methylation abnormalities at imprinting centres) leading to defects in the expression of imprinted genes. The main objective of our team is to improve our knowledge of the pathophysiological and environmental mechanisms involved in the regulation of fetal and postnatal growth. To do this, we benefit from a cross-sectional approach through clinical and fundamental studies based on cohorts.
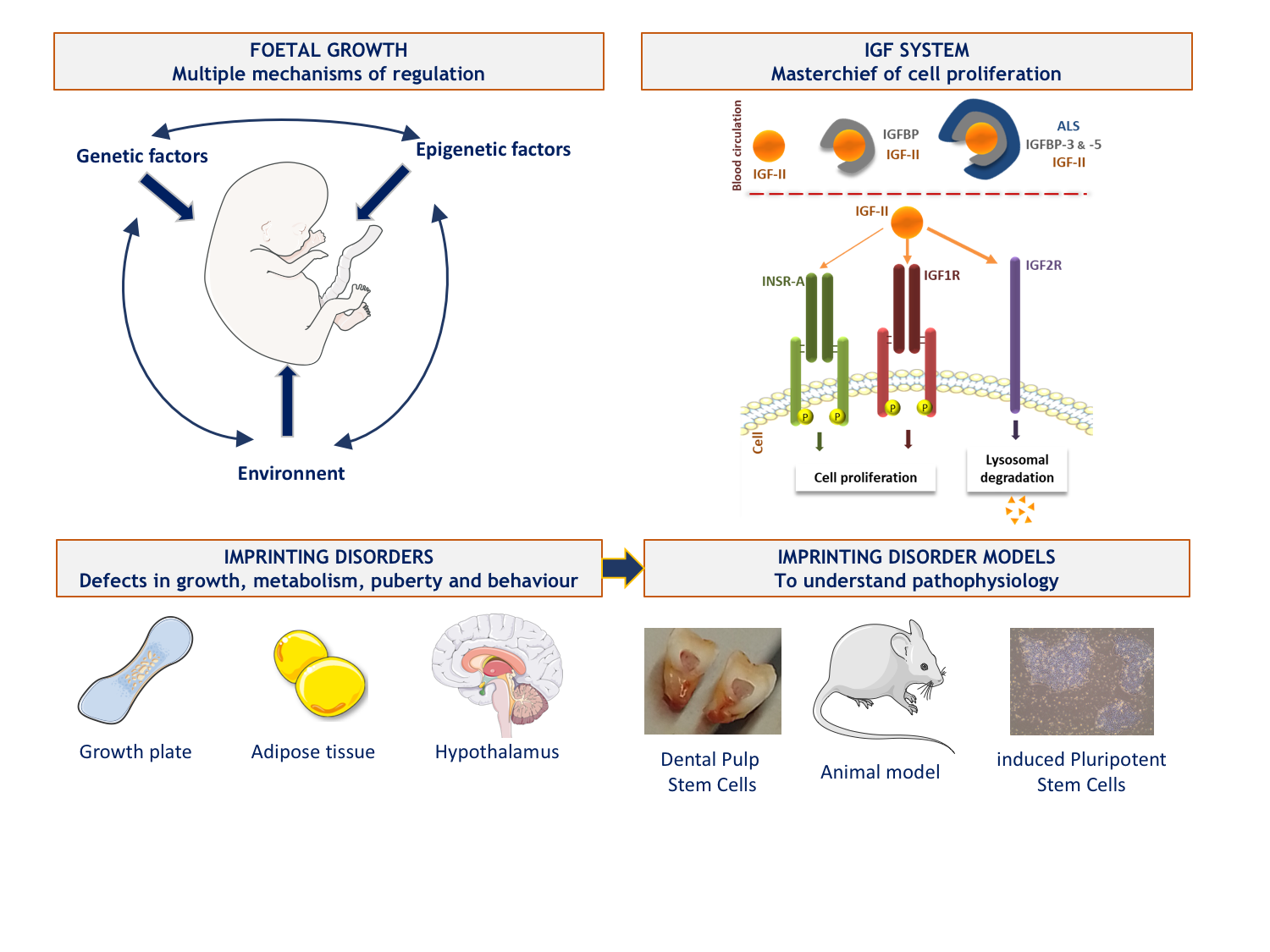
2. Recent and current work
A/ We have developed a high-throughput sequencing panel of genes involved in fetal growth restriction or excess in order to improve the diagnosis of genetic abnormalities in patients with fetal growth defects in France. In addition to genes already described in syndromes close to SRS and BWS, we have included some genes of interest for which no abnormality has been identified in humans but which are theoretically involved in the regulation of fetal growth. Most of them are part of the IGF system as their binding proteins or downstream effectors of their common receptor, IRS1 and IRS2.
B/ In the absence of a fully relevant study model to investigate the pathophysiology of imprinted disorders we are interested in, we have developed two main cellular models.
We have a promising model based on dental pulp stem cells - which we can collect when patients undergo scheduled dental extraction - whose multipotency allows differentiation into cells of interest, involved in the phenotype of the patients. This very recent work has been done in parallel with the reprogramming of induced pluripotent stem cells (iPSCs) from leukocytes. We were able to overcome the hypermethylation profile identified for almost all ICRs in these cells - as also reported by other teams - by developing reprogramming culture conditions that allowed the restoration of balanced methylation in some clones. This innovative protocol was the subject of a publication and a patent application (n°22306056.7).
Currently, we are differentiating these two types of stem cells into cells involved in the phenotype of patients such as hypertrophic chondrocytes (key cells of the human growth plate) or adipocytes. Thus, we will be able to characterise the transcriptomic, developmental and functional differences existing in these cells from controls and patients and better understand the pathophysiological consequences of imprinting diseases, as a first step towards the identification and development of new therapeutic targets.
In addition, in the future, we plan to complement the data from these cellular studies with studies in mouse models of imprinting and IGF system disruption through the development of genome and epigenome editing techniques.
C/ On the other hand, in patients with imprinting disorders, methylation defects may extend to other imprinted loci outside the region initially identified as disrupted. This phenomenon is called multilocus imprinting disorder (MLID). Today, the prevalence and clinical consequences of MLID are poorly described. With the help of high-throughput sequencing technologies, we are developing new molecular tools to analyse the methylation of a large number of imprinted loci in a single step.
D/ Finally, our last aspect of research is to study the impact of the environment on imprinting disorders with abnormal fetal growth. The difficulties encountered in obtaining iPSCs with normal methylation at the level of ICRs and the influence of culture conditions echo the increased prevalence of imprinting diseases in children born by assisted reproduction technologies, a fact already documented in the literature and previously studied in our team by a PHRC (Pr Yves Le Bouc). Indeed, the origin of the epigenetic anomalies responsible for the majority of the imprinting-related disorders we are studying is not known, but the critical phases of setting up and maintaining the methylation of the ICRs take place during gametogenesis and in the first post-conceptional days. Environmental events occurring during these periods are therefore interesting to study in order to assess their effect on the regulation of parental imprinting.
Irène Netchine : ORCID 0000-0003-1324-3389

RADICO-IDMET’ cohort for patients with Imprinting Disorders
Marie-Laure Sobrier: ORCID ID 0000-0001-6396-100X
Eloïse Giabicani : ORCID ID 0000-0001-5360-8616
Frédéric Brioude : ORCID 0000-0001-8122-760X
Laurent Kappeler : ORCID 0000-0001-7971-0838
Clémence Girardet: ORCID 0000-0002-0302-3350
Inserm Biblio - Ressources numériques
Perinatal features of children with Silver-Russell syndrome due to 11p15 loss of methylation.
Darneau D, Giabicani E, Netchine I, Pham A. Front Pediatr. 2024 Apr 4;12:1367433. doi: 10.3389/fped.2024.1367433.
Pubertal origin of growth retardation in inborn errors of protein metabolism: A longitudinal cohort study.
Busiah K, Roda C, Crosnier AS, Brassier A, Servais A, Wicker C, Dubois S, Assoun M, Belloche C, Ottolenghi C, Pontoizeau C, Souberbielle JC, Piketty ML, Perin L, Le Bouc Y, Arnoux JB, Netchine I, Imbard A, de Lonlay P. Mol Genet Metab. 2024 Mar;141(3):108123. doi: 10.1016/j.ymgme.2023.108123.
Ciliopathy due to POC1A deficiency: clinical and metabolic features, and cellular modeling.
Perge K, Capel E, Villanueva C, Gautheron J, Diallo S, Auclair M, Rondeau S, Morichon R, Brioude F, Jéru I, Rossi M, Nicolino M, Vigouroux C. Eur J Endocrinol. 2024 Feb 1;190(2):151-164. doi: 10.1093/ejendo/lvae009.
Post-ligation cardiac syndrome after surgical versus transcatheter closure of patent ductus arteriosus in low body weight premature infants: a multicenter retrospective cohort study.
Duboue PM, Padovani P, Bouteiller XP, Martin-Kabore F, Benbrik N, Gronier CG, Bouissou A, Garnier E, Mitanchez D, Flamant C, Rozé JC, Baruteau AE, Lefort B. Eur J Pediatr. 2024 May;183(5):2193-2201. doi: 10.1007/s00431-024-05481-y.
Electronic reporting of rare endocrine conditions within a clinical network: results from the EuRRECa project.
Ali SR, Bryce J, Priego-Zurita AL, Cherenko M, Smythe C, de Rooij TM, Cools M, Danne T, Katugampola H, Dekkers OM, Hiort O, Linglart A, Netchine I, Nordenstrom A, Attila P, Persani L, Reisch N, Smyth A, Sumnik Z, Taruscio D, Visser WE, Pereira AM, Appelman-Dijkstra NM, Ahmed SF. Endocr Connect. 2023 Nov 20;12(12):e230434. doi: 10.1530/EC-23-0434.
https://ec.bioscientifica.com/view/journals/ec/12/12/EC-23-0434.xml
Executive functioning in adolescents and adults with Silver-Russell syndrome.
Burgevin M, Lacroix A, Ollivier F, Bourdet K, Coutant R, Donadille B, Faivre L, Manouvrier-Hanu S, Petit F, Thauvin-Robinet C, Toutain A, Netchine I, Odent S. PLoS One. 2023 Jan 20;18(1):e0279745. doi: 10.1371/journal.pone.0279745.
Imprinting disorders.
Eggermann T, Monk D, de Nanclares GP, Kagami M, Giabicani E, Riccio A, Tümer Z, Kalish JM, Tauber M, Duis J, Weksberg R, Maher ER, Begemann M, Elbracht M. Nat Rev Dis Primers. 2023 Jun 29;9(1):33. doi: 10.1038/s41572-023-00443-4.
Maintenance of methylation profile in imprinting control regions in human induced pluripotent stem cells.
Pham A, Selenou C, Giabicani E, Fontaine V, Marteau S, Brioude F, David L, Mitanchez D, Sobrier ML, Netchine I. Clin Epigenetics. 2022 14(1):190. doi: 10.1186/s13148-022-01410-8.
Dental pulp stem cells as a promising model to study imprinting diseases.
Giabicani E, Pham A, Sélénou C, Sobrier ML, Andrique C, Lesieur J, LinglartA, Poliard A, Chaussain C, Netchine I. Int J Oral Sci. 2022 Apr 2;14(1):19. doi:10.1038/s41368-022-00169-1.
IGF2 : Development, Genetic and Epigenetic Abnormalities.
Sélénou C, Brioude F, Giabicani E, Sobrier ML, Netchine I. Cells. 2022 11(12):1886. doi: 10.3390/cells11121886.
Low Maternal DLK1 Levels at 26 Weeks Is Associated With Small for Gestational Age at Birth.
Pham A, Mitanchez D, Forhan A, Perin L, Le Bouc Y, Brioude F, Sobrier ML,Heude B, Netchine I. Front Endocrinol (Lausanne). 2022 13:836731. doi: 10.3389/fendo.2022.836731.
Increasing knowledge in IGF1R defects: lessons from 35 new patients.
Giabicani E, Willems M, Steunou V, Chantot-Bastaraud S, Thibaud N, Abi Habib W, Azzi S, Lam B, Bérard L, Bony-Trifunovic H, Brachet C, Brischoux-Boucher E, Caldagues E, Coutant R, Cuvelier ML, Gelwane G, Guemas I, Houang M, Isidor B, Jeandel C, Lespinasse J, Naud-Saudreau C, Jesuran-Perelroizen M, Perrin L, Piard J, Sechter C, Souchon PF, Storey C, Thomas D, Le Bouc Y, Rossignol S, Netchine I, Brioude F. J Med Genet. 2020 57(3):160-168. doi:10.1136/jmedgenet-2019-106328.
Overgrowth syndromes - clinical and molecular aspects and tumour risk.
Brioude F, Toutain A, Giabicani E, Cottereau E, Cormier-Daire V, Netchine I. Nat Rev Endocrinol. 2019 15(5):299-311. doi: 10.1038/s41574-019-0180-z.
Transcriptional profiling at the DLK1/MEG3 domain explains clinical overlap between imprinting disorders.
Abi Habib W, Brioude F, Azzi S, Rossignol S, Linglart A, Sobrier ML, Giabicani É, Steunou V, Harbison MD, Le Bouc Y, Netchine I. Sci Adv. 2019 5(2):eaau9425. doi:10.1126/sciadv.aau9425.
Genetic disruption of the oncogenic HMGA2-PLAG1-IGF2 pathway causes fetal growth restriction.
Abi Habib W, Brioude F, Edouard T, Bennett JT, Lienhardt-Roussie A, Tixier F,Salem J, Yuen T, Azzi S, Le Bouc Y, Harbison MD, Netchine I. Genet Med. 2018 20(2):250-258. doi: 10.1038/gim.2017.105
Chromosome 14q32.2 Imprinted Region Disruption as an Alternative Molecular Diagnosis of Silver-Russell Syndrome.
Geoffron S, Abi Habib W, Chantot-Bastaraud S, Dubern B, Steunou V, Azzi S, Afenjar A, Busa T, Pinheiro Canton A, Chalouhi C, Dufourg MN, Esteva B, Fradin M, Geneviève D, Heide S, Isidor B, Linglart A, Morice Picard F, Naud-Saudreau C, Oliver Petit I, Philip N, Pienkowski C, Rio M, Rossignol S, Tauber M, Thevenon J, Vu-Hong TA, Harbison MD, Salem J, Brioude F, Netchine I, Giabicani E. J Clin Endocrinol Metab. 2018 103(7):2436-2446. doi: 10.1210/jc.2017-02152. PMID: 29659920
Diagnosis and management of Silver-Russell syndrome: first international consensus statement.
Wakeling EL, Brioude F, Lokulo-Sodipe O, O'Connell SM, Salem J, Bliek J,Canton AP, Chrzanowska KH, Davies JH, Dias RP, Dubern B, Elbracht M, Giabicani E, Grimberg A, Grønskov K, Hokken-Koelega AC, Jorge AA, Kagami M, Linglart A,Maghnie M, Mohnike K, Monk D, Moore GE, Murray PG, Ogata T, Petit IO, Russo S,Said E, Toumba M, Tümer Z, Binder G, Eggermann T, Harbison MD, Temple IK, MackayDJ, Netchine I. Nat Rev Endocrinol. 2017 13(2):105-124. doi: 10.1038/nrendo.2016.138.
INSERM
Kourilsky Building
34 rue Crozatier - 75012 Paris
France
Sorbonne Université Medicine
Saint-Antoine Site
27 rue Chaligny - 75012 Paris
France
